This article was published as a part of the Data Science Blogathon
The ongoing global pandemic called COVID-19 is caused by SARS-COV-2, which was first found and identified in Wuhan, China, in December 2019. The lockdown in Wuhan and its neighboring districts failed to contain this virus which led to one of the worst humanitarian crises in the modern world, affecting millions of people all around the globe.
The virus spread quickly and mutated which gave rise to several waves of it, mostly affecting the third world and developing countries. The number of affected people is rising steadily as the world’s governments try to control the spread.

In this article, we will use the CoronaHack- Chest X-Ray Dataset. It contains chest X-Ray images and we have to find the ones that are affected by the coronavirus.
The SARS-COV-2 which we talked previously about is the type of virus that majorly affects the respiratory system, so Chest X-ray is one of the important imaging methods we can use to identify an affected lung. Here’s a side by side comparison:
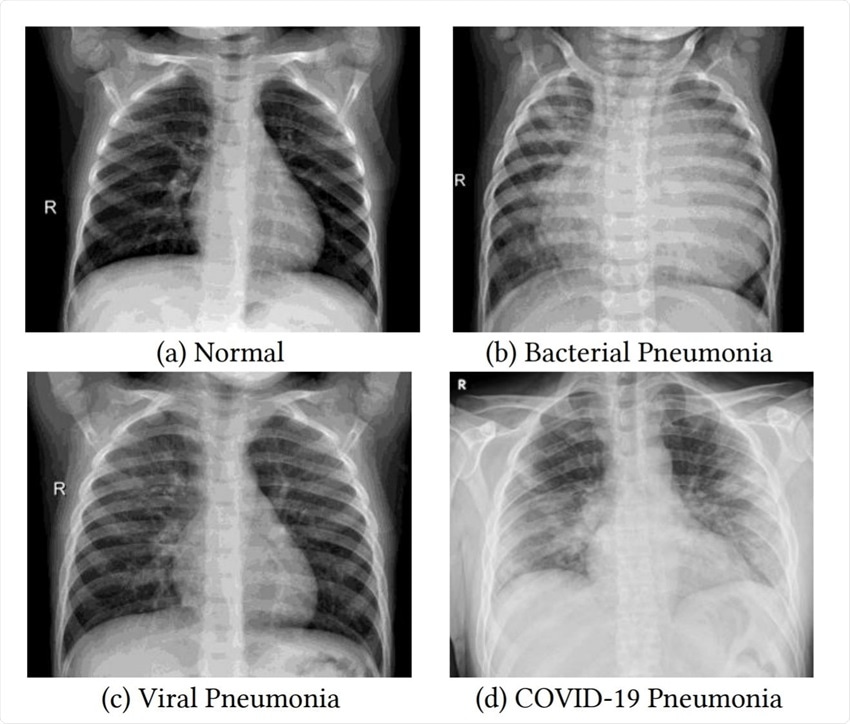
So as you can see, how COVID-19 pneumonia can engulf the whole lungs and is more dangerous than both Bacterial as well as Viral types of pneumonia. I highly suggest you read the paper Transfer Learning for COVID-19 Pneumonia Detection
and Classification in Chest X-ray Images which I’ve linked in the above image.
In this article, we will use Deep learning and Transfer learning to classify and identify X-Ray Images of lungs affected with Covid-19.
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
import numpy as np
import pandas as pd
sns.set()
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import *
from tensorflow.keras.optimizers import Adam, SGD, RMSprop
from tensorflow.keras.applications import DenseNet121, VGG19, ResNet50
import PIL.Image
import matplotlib.pyplot as mpimg
import os
from tensorflow.keras.preprocessing.image import ImageDataGenerator, img_to_array
from tensorflow.keras.preprocessing import image
from tqdm import tqdm
import warnings
warnings.filterwarnings("ignore")
from sklearn.utils import shuffle
train_df = pd.read_csv('../input/coronahack-chest-xraydataset/Chest_xray_Corona_Metadata.csv')
train_df.shape
> (5910, 6)
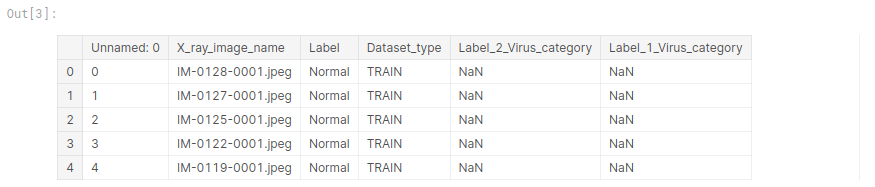
train_df.head(5)
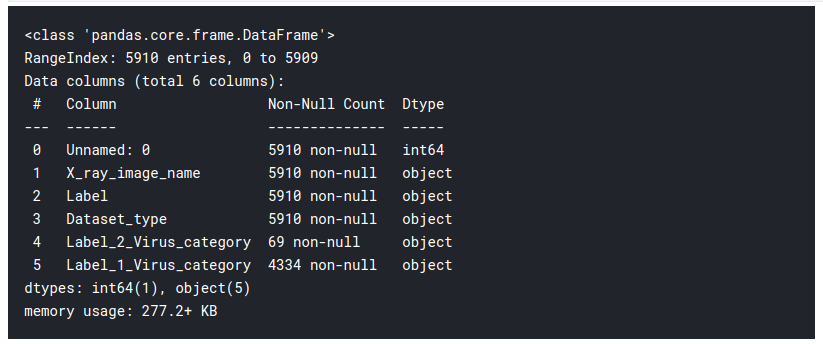
train_df.info()
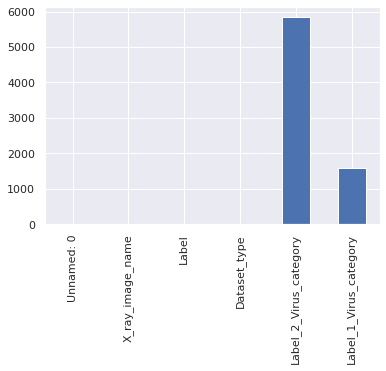
missing_vals = train_df.isnull().sum() missing_vals.plot(kind = 'bar')
train_df.dropna(how = 'all') train_df.isnull().sum()

train_df.fillna('unknown', inplace=True)
train_df.isnull().sum()

train_data = train_df[train_df['Dataset_type'] == 'TRAIN']
test_data = train_df[train_df['Dataset_type'] == 'TEST']
assert train_data.shape[0] + test_data.shape[0] == train_df.shape[0]
print(f"Shape of train data : {train_data.shape}")
print(f"Shape of test data : {test_data.shape}")
test_data.sample(10)

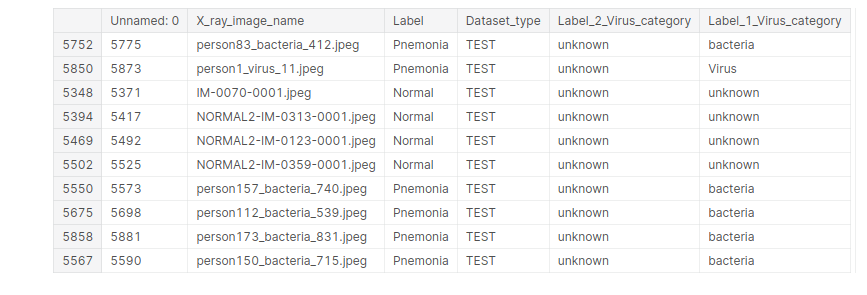
We will fill the missing values with ‘unknown’.
print((train_df['Label_1_Virus_category']).value_counts())
print('--------------------------')
print((train_df['Label_2_Virus_category']).value_counts())
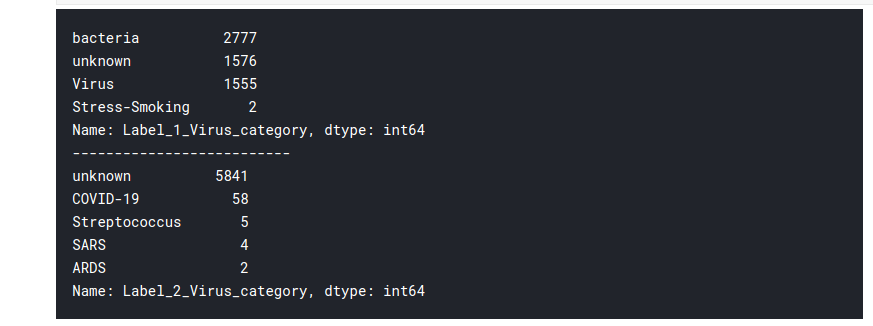
Thus the Label 2 category contains the COVID-19 cases!
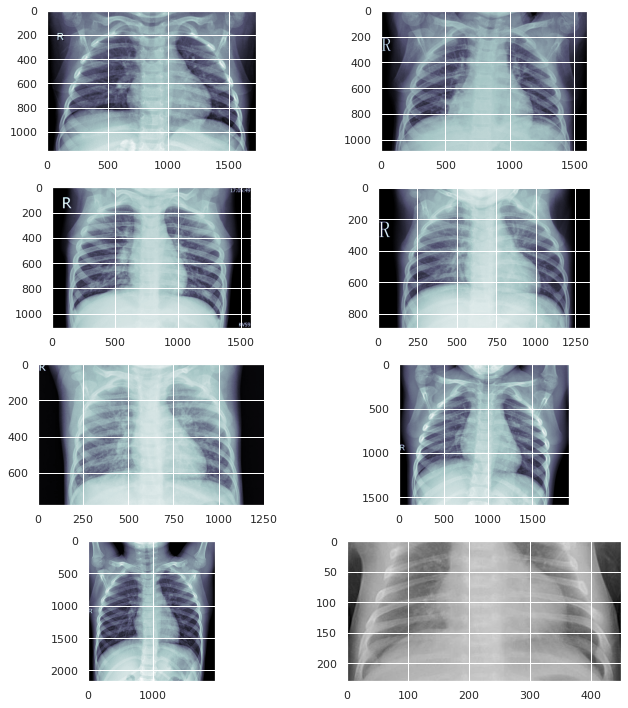
test_img_dir = '/kaggle/input/coronahack-chest-xraydataset/Coronahack-Chest-XRay-Dataset/Coronahack-Chest-XRay-Dataset/test' train_img_dir = '/kaggle/input/coronahack-chest-xraydataset/Coronahack-Chest-XRay-Dataset/Coronahack-Chest-XRay-Dataset/train' sample_train_images = list(os.walk(train_img_dir))[0][2][:8] sample_train_images = list(map(lambda x: os.path.join(train_img_dir, x), sample_train_images)) sample_test_images = list(os.walk(test_img_dir))[0][2][:8] sample_test_images = list(map(lambda x: os.path.join(test_img_dir, x), sample_test_images))
plt.figure(figsize = (10,10))
for iterator, filename in enumerate(sample_train_images):
image = PIL.Image.open(filename)
plt.subplot(4,2,iterator+1)
plt.imshow(image, cmap=plt.cm.bone)
plt.tight_layout()
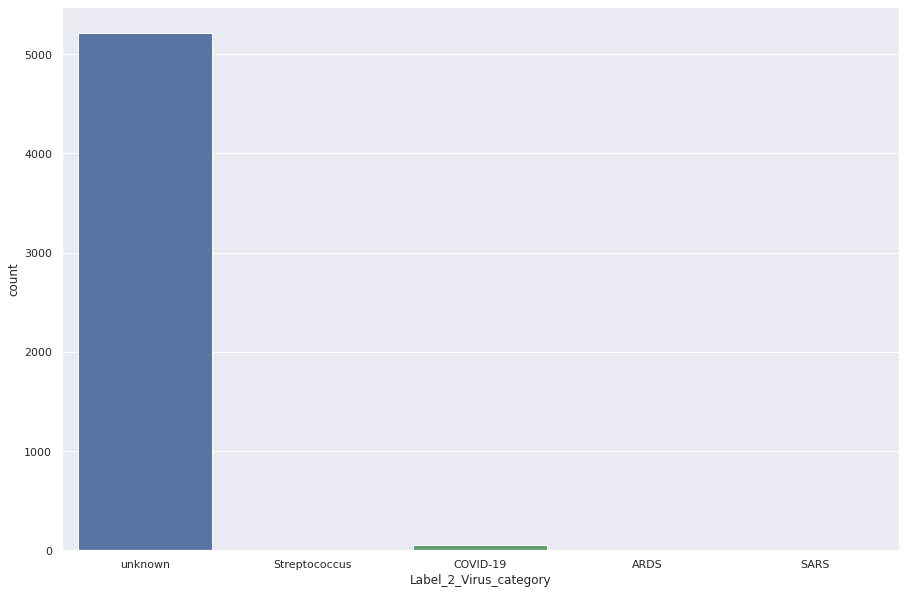
plt.figure(figsize=(15,10)) sns.countplot(train_data['Label_2_Virus_category']);
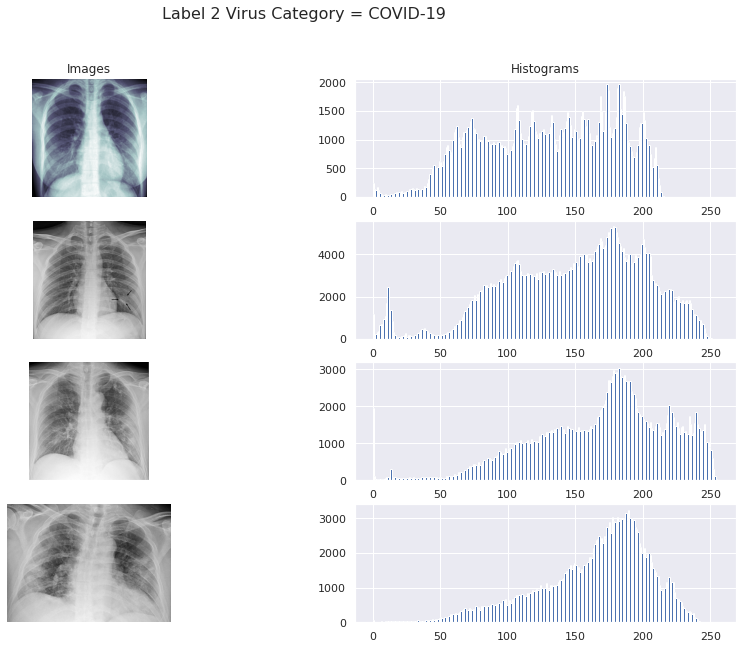
For COVID-19 cases
fig, ax = plt.subplots(4, 2, figsize=(15, 10))
covid_path = train_data[train_data['Label_2_Virus_category']=='COVID-19']['X_ray_image_name'].values
sample_covid_path = covid_path[:4]
sample_covid_path = list(map(lambda x: os.path.join(train_img_dir, x), sample_covid_path))
for row, file in enumerate(sample_covid_path):
image = plt.imread(file)
ax[row, 0].imshow(image, cmap=plt.cm.bone)
ax[row, 1].hist(image.ravel(), 256, [0,256])
ax[row, 0].axis('off')
if row == 0:
ax[row, 0].set_title('Images')
ax[row, 1].set_title('Histograms')
fig.suptitle('Label 2 Virus Category = COVID-19', size=16)
plt.show()
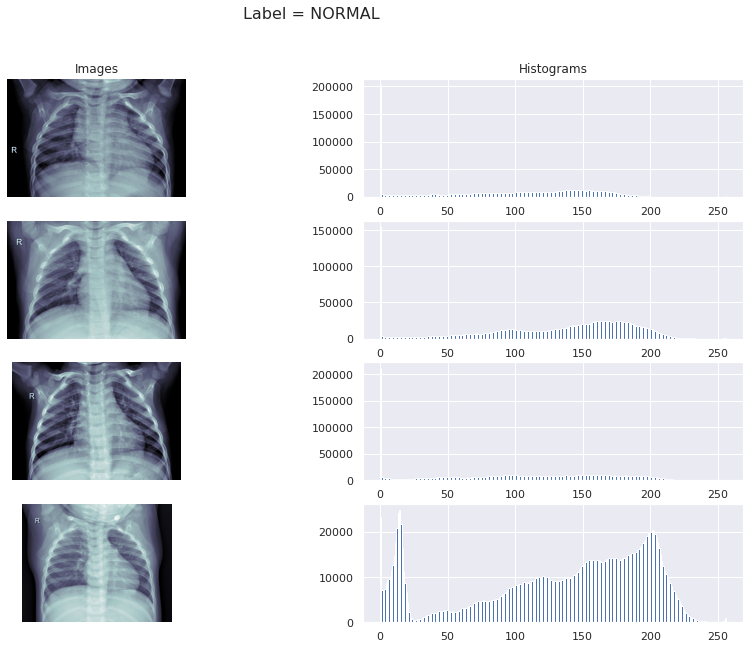
For Normal cases
fig, ax = plt.subplots(4, 2, figsize=(15, 10))
normal_path = train_data[train_data['Label']=='Normal']['X_ray_image_name'].values
sample_normal_path = normal_path[:4]
sample_normal_path = list(map(lambda x: os.path.join(train_img_dir, x), sample_normal_path))
for row, file in enumerate(sample_normal_path):
image = plt.imread(file)
ax[row, 0].imshow(image, cmap=plt.cm.bone)
ax[row, 1].hist(image.ravel(), 256, [0,256])
ax[row, 0].axis('off')
if row == 0:
ax[row, 0].set_title('Images')
ax[row, 1].set_title('Histograms')
fig.suptitle('Label = NORMAL', size=16)
plt.show()
final_train_data = train_data[(train_data['Label'] == 'Normal') |
((train_data['Label'] == 'Pnemonia') &
(train_data['Label_2_Virus_category'] == 'COVID-19'))]
final_train_data['class'] = final_train_data.Label.apply(lambda x: 'negative' if x=='Normal' else 'positive') test_data['class'] = test_data.Label.apply(lambda x: 'negative' if x=='Normal' else 'positive') final_train_data['target'] = final_train_data.Label.apply(lambda x: 0 if x=='Normal' else 1) test_data['target'] = test_data.Label.apply(lambda x: 0 if x=='Normal' else 1)
final_train_data = final_train_data[['X_ray_image_name', 'class', 'target', 'Label_2_Virus_category']] final_test_data = test_data[['X_ray_image_name', 'class', 'target']]
test_data['Label'].value_counts()
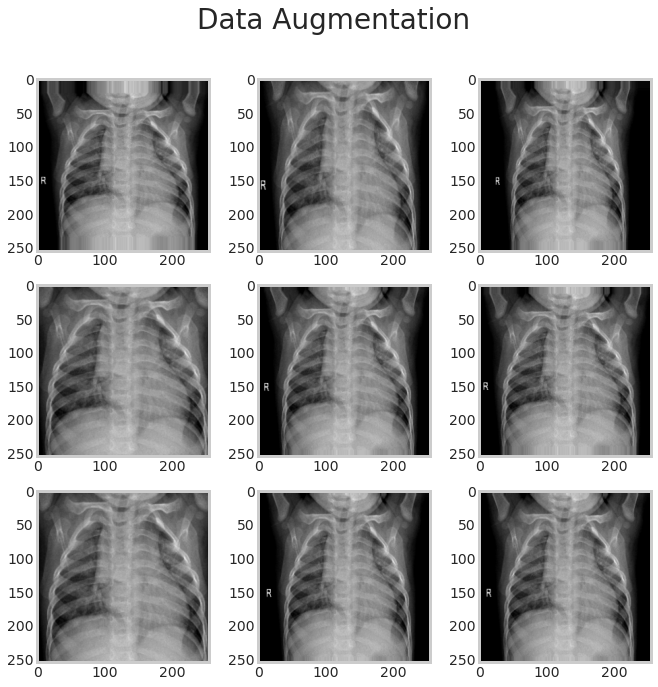
datagen = ImageDataGenerator(
shear_range=0.2,
zoom_range=0.2,
)
def read_img(filename, size, path):
img = image.load_img(os.path.join(path, filename), target_size=size)
#convert image to array
img = image.img_to_array(img) / 255
return img
samp_img = read_img(final_train_data['X_ray_image_name'][0],
(255,255),
train_img_path)
plt.figure(figsize=(10,10))
plt.suptitle('Data Augmentation', fontsize=28)
i = 0
for batch in datagen.flow(tf.expand_dims(samp_img,0), batch_size=6):
plt.subplot(3, 3, i+1)
plt.grid(False)
plt.imshow(batch.reshape(255, 255, 3));
if i == 8:
break
i += 1
plt.show();
corona_df = final_train_data[final_train_data['Label_2_Virus_category'] == 'COVID-19']
with_corona_augmented = []
def augment(name):
img = read_img(name, (255,255), train_img_path)
i = 0
for batch in tqdm(datagen.flow(tf.expand_dims(img, 0), batch_size=32)):
with_corona_augmented.append(tf.squeeze(batch).numpy())
if i == 20:
break
i =i+1
corona_df['X_ray_image_name'].apply(augment)
Note: The output was too long to include in the article. Here’s a small part of it.

train_arrays = [] final_train_data['X_ray_image_name'].apply(lambda x: train_arrays.append(read_img(x, (255,255), train_img_dir))) test_arrays = [] final_test_data['X_ray_image_name'].apply(lambda x: test_arrays.append(read_img(x, (255,255), test_img_dir)))
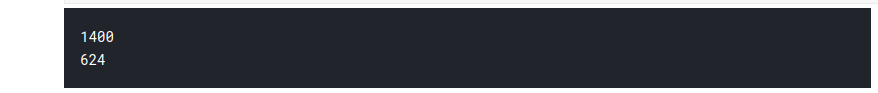
print(len(train_arrays)) print(len(test_arrays))
y_train = np.concatenate((np.int64(final_train_data['target'].values), np.ones(len(with_corona_augmented), dtype=np.int64)))
train_tensors = tf.convert_to_tensor(np.concatenate((np.array(train_arrays), np.array(with_corona_augmented)))) test_tensors = tf.convert_to_tensor(np.array(test_arrays)) y_train_tensor = tf.convert_to_tensor(y_train) y_test_tensor = tf.convert_to_tensor(final_test_data['target'].values) train_dataset = tf.data.Dataset.from_tensor_slices((train_tensors, y_train_tensor)) test_dataset = tf.data.Dataset.from_tensor_slices((test_tensors, y_test_tensor))
for i,l in train_dataset.take(1):
plt.imshow(i);
BATCH_SIZE = 16
BUFFER = 1000
train_batches = train_dataset.shuffle(BUFFER).batch(BATCH_SIZE)
test_batches = test_dataset.batch(BATCH_SIZE)
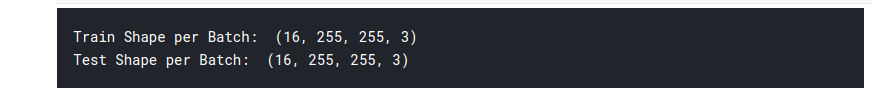
for i,l in train_batches.take(1):
print('Train Shape per Batch: ',i.shape);
for i,l in test_batches.take(1):
print('Test Shape per Batch: ',i.shape);
INPUT_SHAPE = (255,255,3)
base_model = tf.keras.applications.ResNet50(input_shape= INPUT_SHAPE,
include_top=False,
weights='imagenet')
# We set it to False because we don't want to mess with the pretrained weights of the model.
base_model.trainable = False
Now our Transfer Learning is successful!!

for i,l in train_batches.take(1):
pass
base_model(i).shape
> TensorShape([16, 8, 8, 2048])
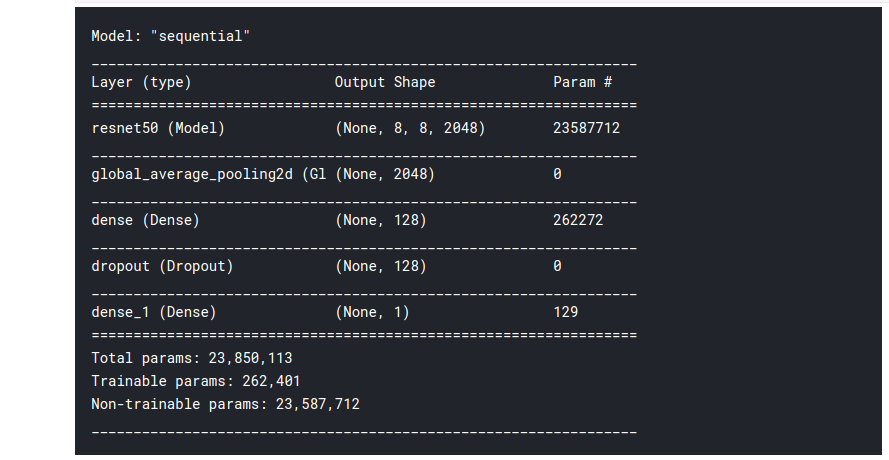
model = Sequential() model.add(base_model) model.add(Layers.GlobalAveragePooling2D()) model.add(Layers.Dense(128)) model.add(Layers.Dropout(0.2)) model.add(Layers.Dense(1, activation = 'sigmoid')) model.summary()
callbacks = tf.keras.callbacks.EarlyStopping(monitor='val_loss', patience=2)
model.compile(optimizer='adam',
loss = 'binary_crossentropy',
metrics=['accuracy'])
model.fit(train_batches, epochs=10, validation_data=test_batches, callbacks=[callbacks])
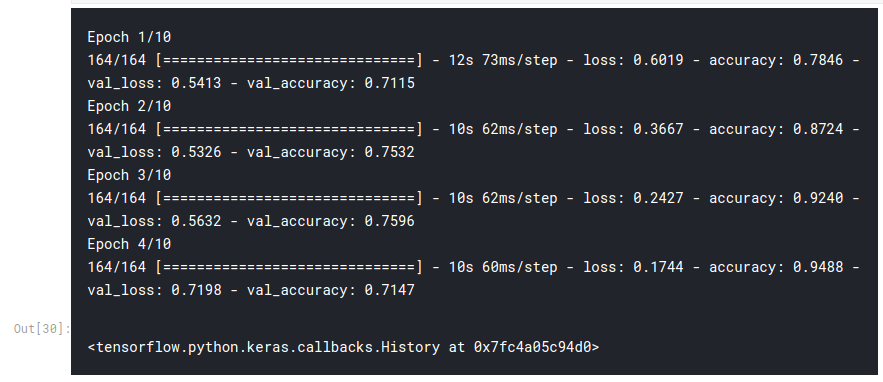
pred = model.predict_classes(np.array(test_arrays))
# classification report from sklearn.metrics import classification_report, confusion_matrix print(classification_report(test_data['target'], pred.flatten()))
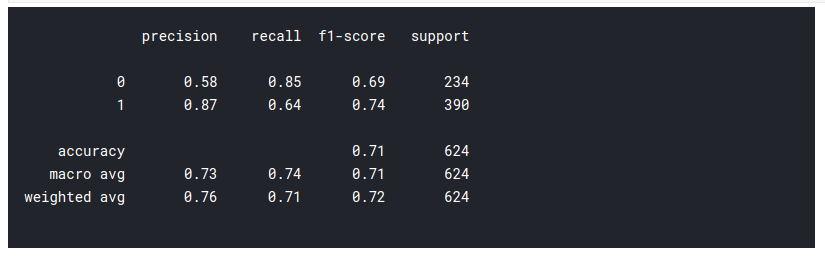
So as you can see the prediction is not half bad. We will plot a confusion matrix to visualize the performance of our model:
con_mat = confusion_matrix(test_data['target'], pred.flatten())
plt.figure(figsize = (10,10))
plt.title('CONFUSION MATRIX')
sns.heatmap(con_mat, cmap='cividis',
yticklabels=['Negative', 'Positive'],
xticklabels=['Negative', 'Positive'],
annot=True);
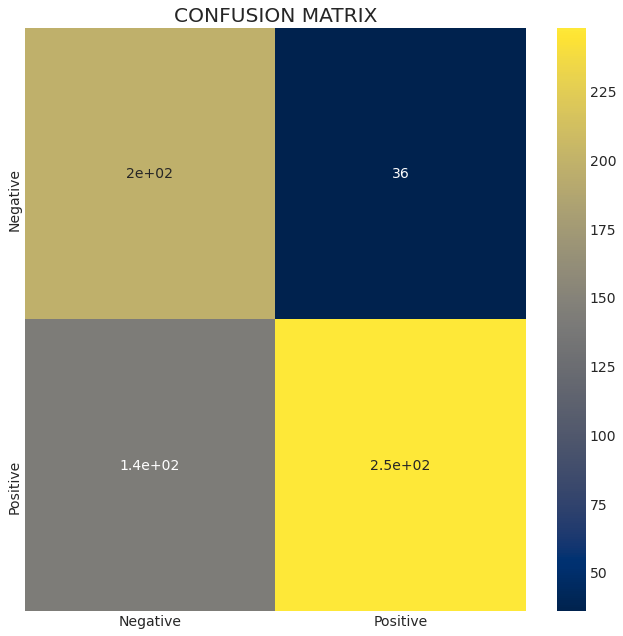
This dataset was an interesting one, and the more I am learning Data Science and Machine Learning, the more I find this subject interesting. There’s soo many ways we can use data nowadays and using it can save countless lives. Thank you for reading this article. You can read more of my content at:
Lorem ipsum dolor sit amet, consectetur adipiscing elit,
Great. How is transfer learning different from deep